
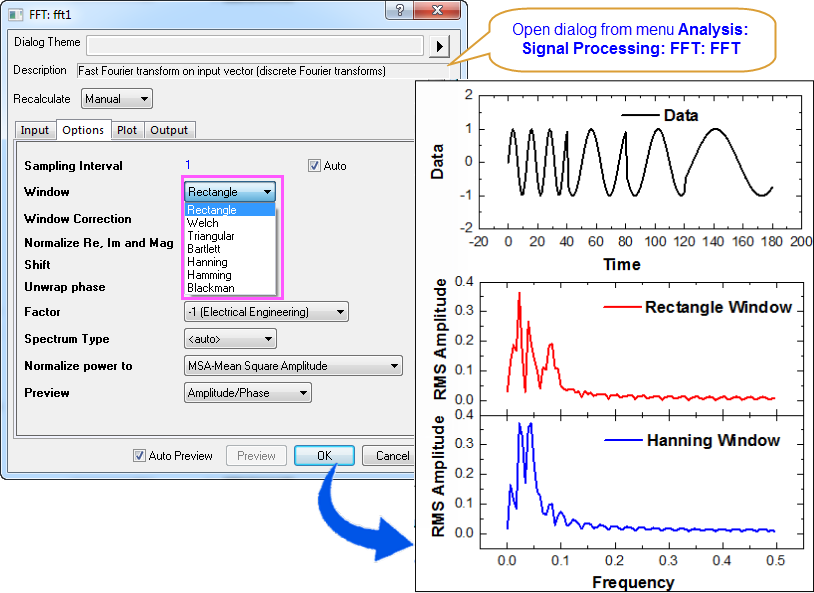
Circular DNA of 3T6R50 double minute chromosomes. Minute chromatin bodies in malignant tumours of childhood. Extrachromosomal oncogene amplification in tumour pathogenesis and evolution. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Extrachromosomal DNA is associated with oncogene amplification and poor outcome across multiple cancers. Extrachromosomal DNA-relieving heredity constraints, accelerating tumour evolution. The role of 3D genome organization in development and cell differentiation. The 3D genome in transcriptional regulation and pluripotency. Circular ecDNA promotes accessible chromatin and high oncogene expression. Thus, protein-tethered ecDNA hubs enable intermolecular transcriptional regulation and may serve as units of oncogene function and cooperative evolution and as potential targets for cancer therapy. Systematic silencing of ecDNA enhancers by CRISPR interference reveals intermolecular enhancer–gene activation among multiple oncogene loci that are amplified on distinct ecDNAs. Furthermore, the PVT1 promoter on an exogenous episome suffices to mediate gene activation in trans by ecDNA hubs in a JQ1-sensitive manner. The BRD4-bound PVT1 promoter is ectopically fused to MYC and duplicated in ecDNA, receiving promiscuous enhancer input to drive potent expression of MYC. The BET inhibitor JQ1 disperses ecDNA hubs and preferentially inhibits ecDNA-derived-oncogene transcription. ecDNA hubs are tethered by the bromodomain and extraterminal domain (BET) protein BRD4 in a MYC-amplified colorectal cancer cell line.
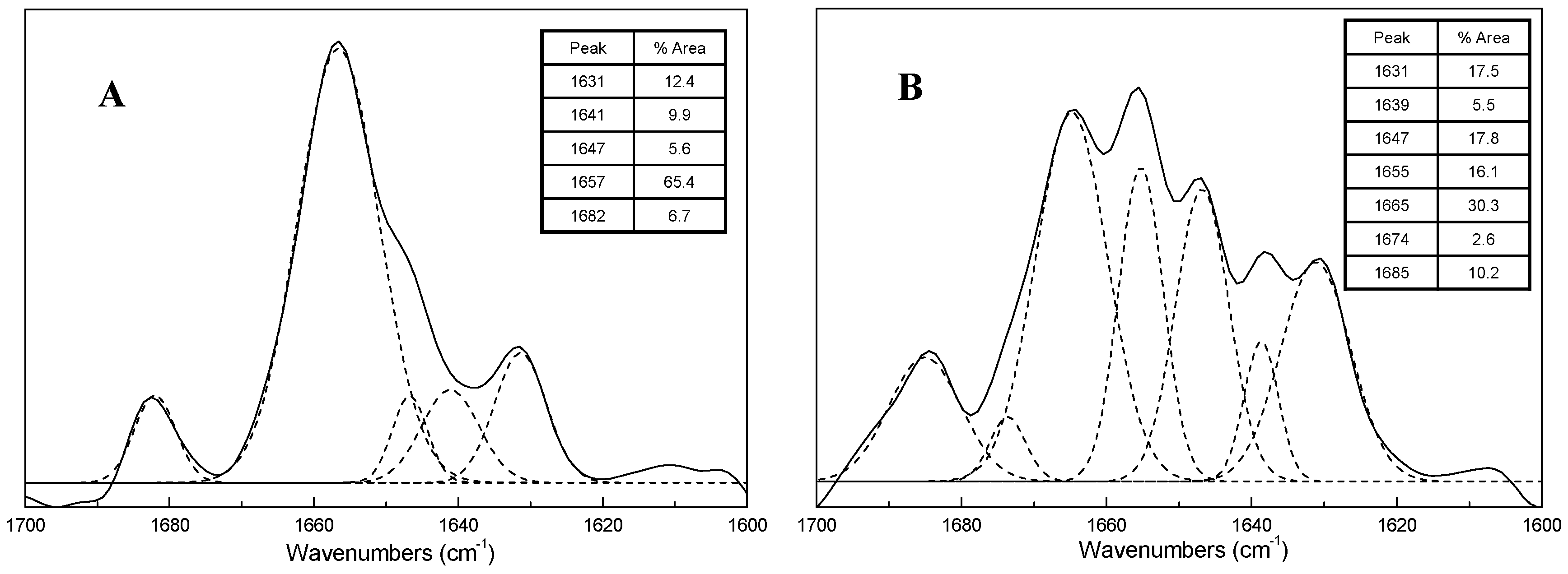
Each ecDNA is more likely to transcribe the oncogene when spatially clustered with additional ecDNAs. ecDNAs that encode multiple distinct oncogenes form hubs in diverse cancer cell types and primary tumours. Here we show that ecDNA hubs-clusters of around 10–100 ecDNAs within the nucleus-enable intermolecular enhancer–gene interactions to promote oncogene overexpression. Gene induction typically involves cis-regulatory elements that contact and activate genes on the same chromosome 2, 3.
Chang ORCID: /0000-0002-9459-4393 1, 21Įxtrachromosomal DNA (ecDNA) is prevalent in human cancers and mediates high expression of oncogenes through gene amplification and altered gene regulation 1. EcDNA hubs drive cooperative intermolecular oncogene expression


 0 kommentar(er)
0 kommentar(er)
